We propose ProtLLM, a versatile cross-modal large language model (LLM) for both protein-centric and protein-language tasks. ProtLLM features a unique dynamic protein mounting mechanism, enabling it to handle complex inputs where the natural language text is interspersed with an arbitrary number of proteins. Besides, we propose the protein-as-word language modeling approach to train ProtLLM. By developing a specialized protein vocabulary, we equip the model with the capability to predict not just natural language but also proteins from a vast pool of candidates. Additionally, we construct a large-scale interleaved protein-text dataset, named InterPT, for pre-training. This dataset comprehensively encompasses both (1) structured data sources like protein annotations and (2) unstructured data sources like biological research papers, thereby endowing ProtLLM with crucial knowledge for understanding proteins. We evaluate ProtLLM on classic supervised protein-centric tasks and explore its novel protein-language applications. Experimental results demonstrate that ProtLLM not only achieves superior performance against protein-specialized baselines on protein-centric tasks but also induces zero-shot and in-context learning capabilities on protein-language tasks.
The architecture of ProtLLM consists of an autoregressive transformer, a protein encoder, and cross-modal connectors. The connectors are placed at the input layer and the output layer of the LLM, respectively. Specifically, the output-layer connector also serves as a prediction head, allowing our model to perform protein retrieval and multi-choice protein answering tasks without requiring the LLM to generate complicated protein names. With dynamic protein mounting, ProtLLM adeptly handles free-form interleaved protein-text sequences with an arbitrary number of proteins in the input. ProtLLM is pre-trained with protein-as-word language modeling that unifies word and protein prediction by constructing a protein vocabulary.
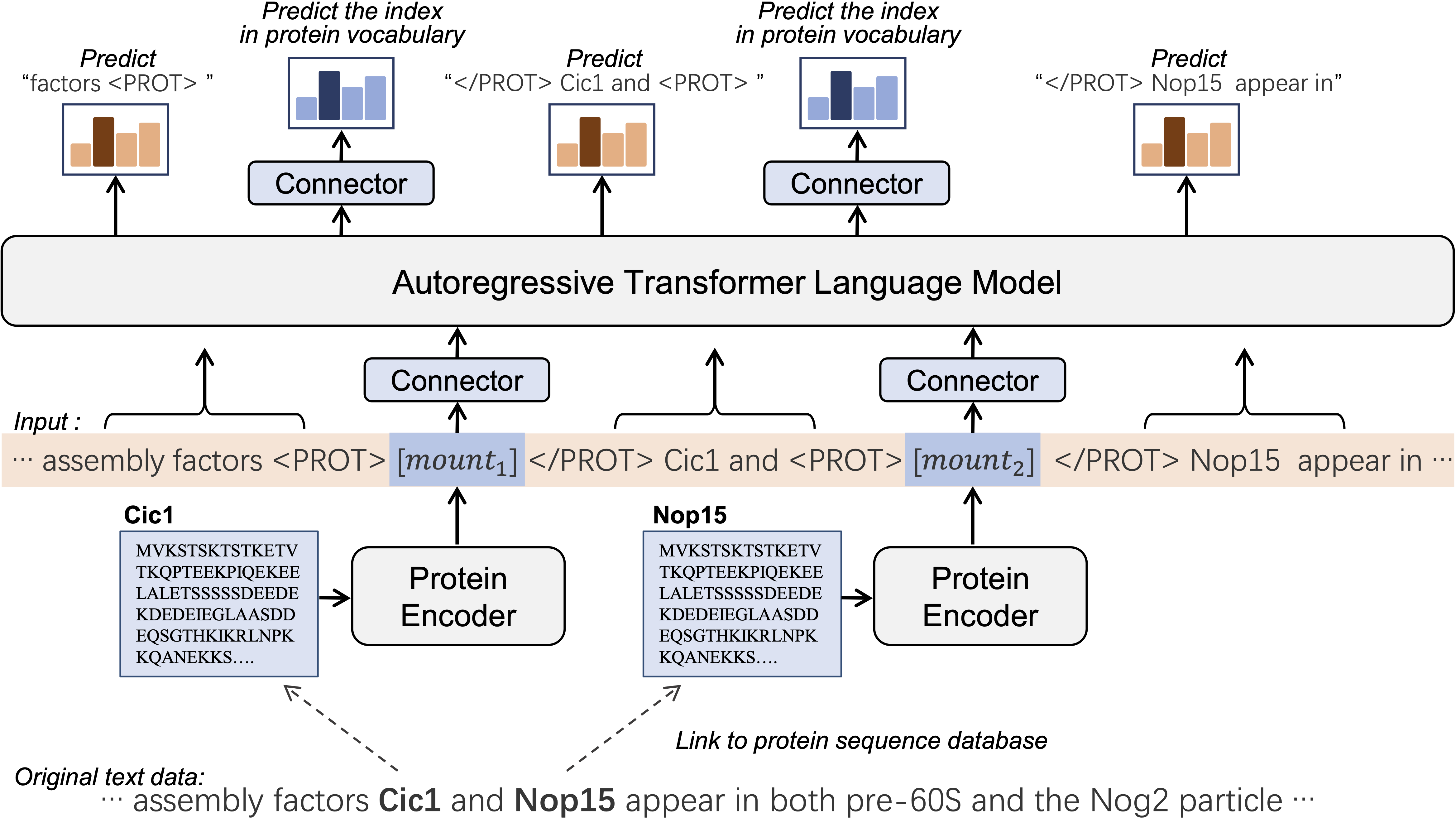
Overview of ProtLLM.
We propose a large-scale interleaved protein-text multimodal dataset, named InterPT, to pre-train ProtLLM with comprehensive protein-related knowledge. This dataset encompasses three types of data sources, i.e., multi-protein scientific articles, protein-annotation pairs, and protein instruction-following data.
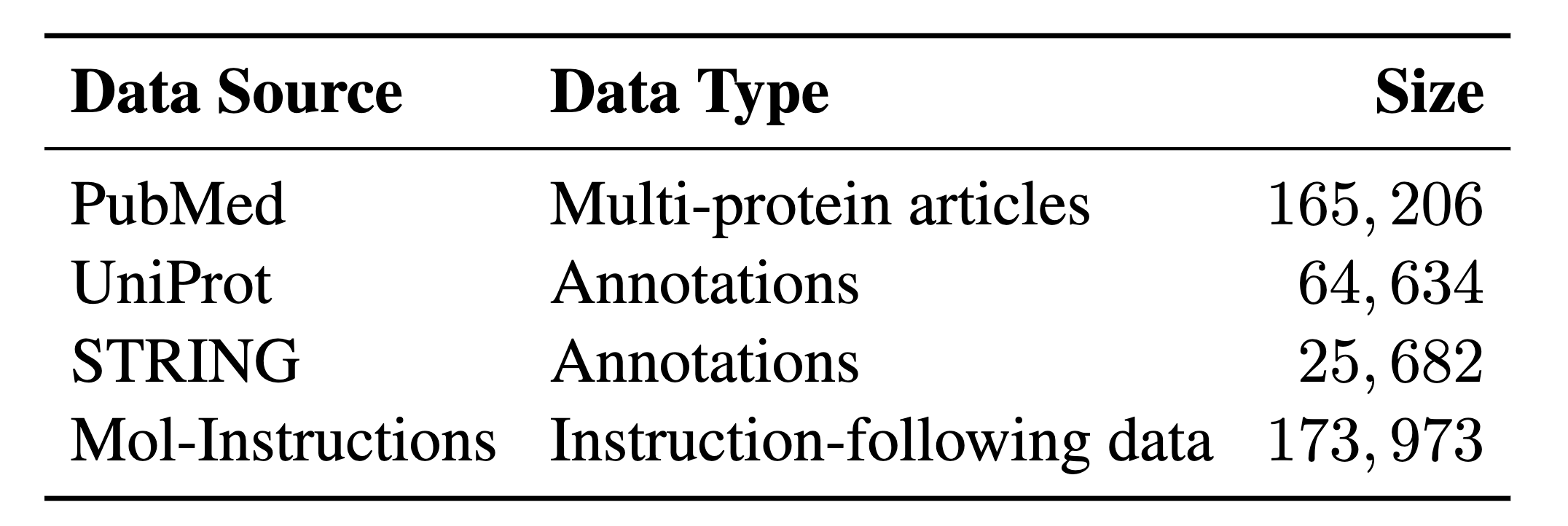
Category and statistics of InterPT components.
We adopt three standard tasks in protein understanding to validate our method: Enzyme Commission (EC) number prediction, Gene Ontology (GO) term prediction, and Protein-Protein Interaction (PPI) prediction. ProtLLM consistently shows competitive or even superior performance compared to both protein-only and protein-text approaches across all benchmarks, indicating the effectiveness of our proposed framework on conventional close-ended protein understanding tasks. Remarkably, ProtLLM obtain 0.596 Fmax and 0.469 AUPR on GO-CC, which outperforms ProtST by a large margin. ProtLLM directly uses pre-trained ProtST as the protein encoder, with the key difference lying in our LLM decoder and pre-training stage for alignment. By comparing ProtLLM with ProtST, the overall improvements strongly highlight the potential benefits of incorporating richer protein-text information and scaling the size of the language model.
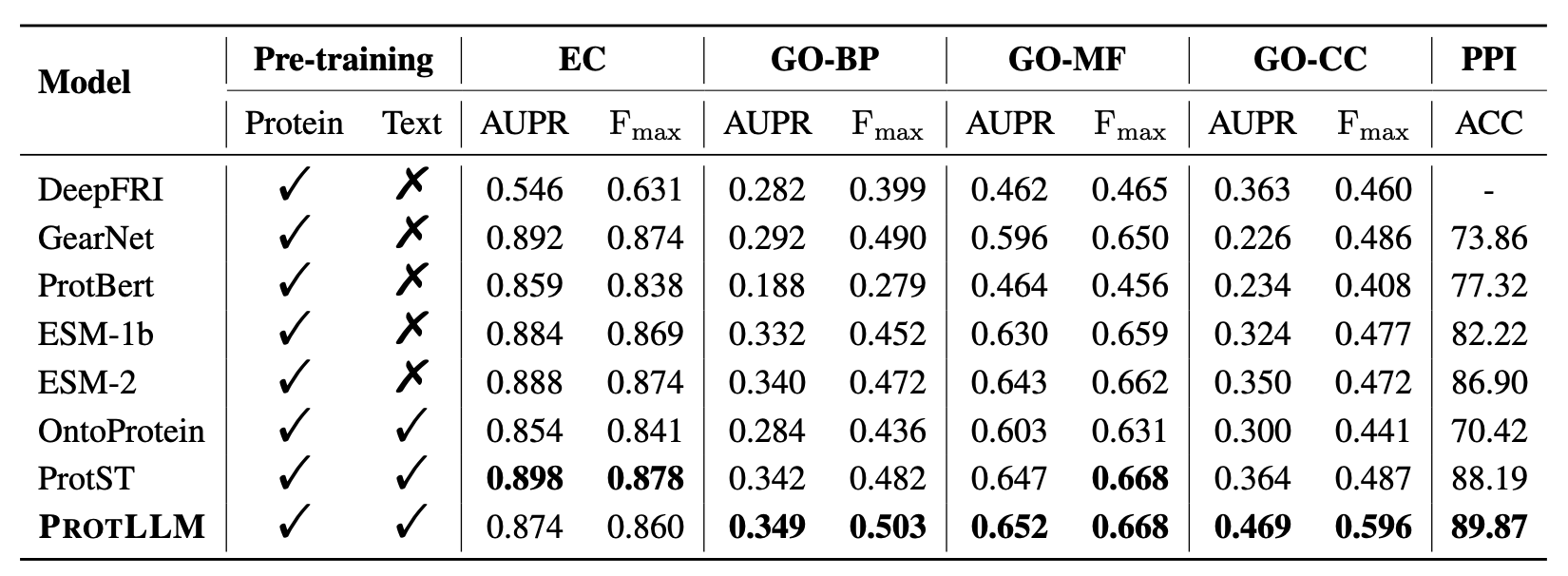
Comparative benchmark results on protein-centric tasks. We use AUPR and Fmax on EC and GO prediction and accuracy (%) on PPI prediction. Bold figures denote the best performance. '-' indicates not applicable.
We investigate whether ProtLLM can achieve in-context learning on the human PPI prediction task. The following figure presents the in-context learning performance on human PPI with varying numbers of demonstration examples. Our model consistently achieves higher PPI accuracy with an increasing number of demonstration examples, demonstrating its effective in-context learning capability for protein-centric tasks. In comparison, the model performs drastically worse upon removing the multi-protein scientific articles, and fails to learn in context with the 2, 6, and 12 demonstrations. We believe that the in-context learning capability of our model could empower biologists to apply it to specialized tasks that lack annotated data, using minimal examples.
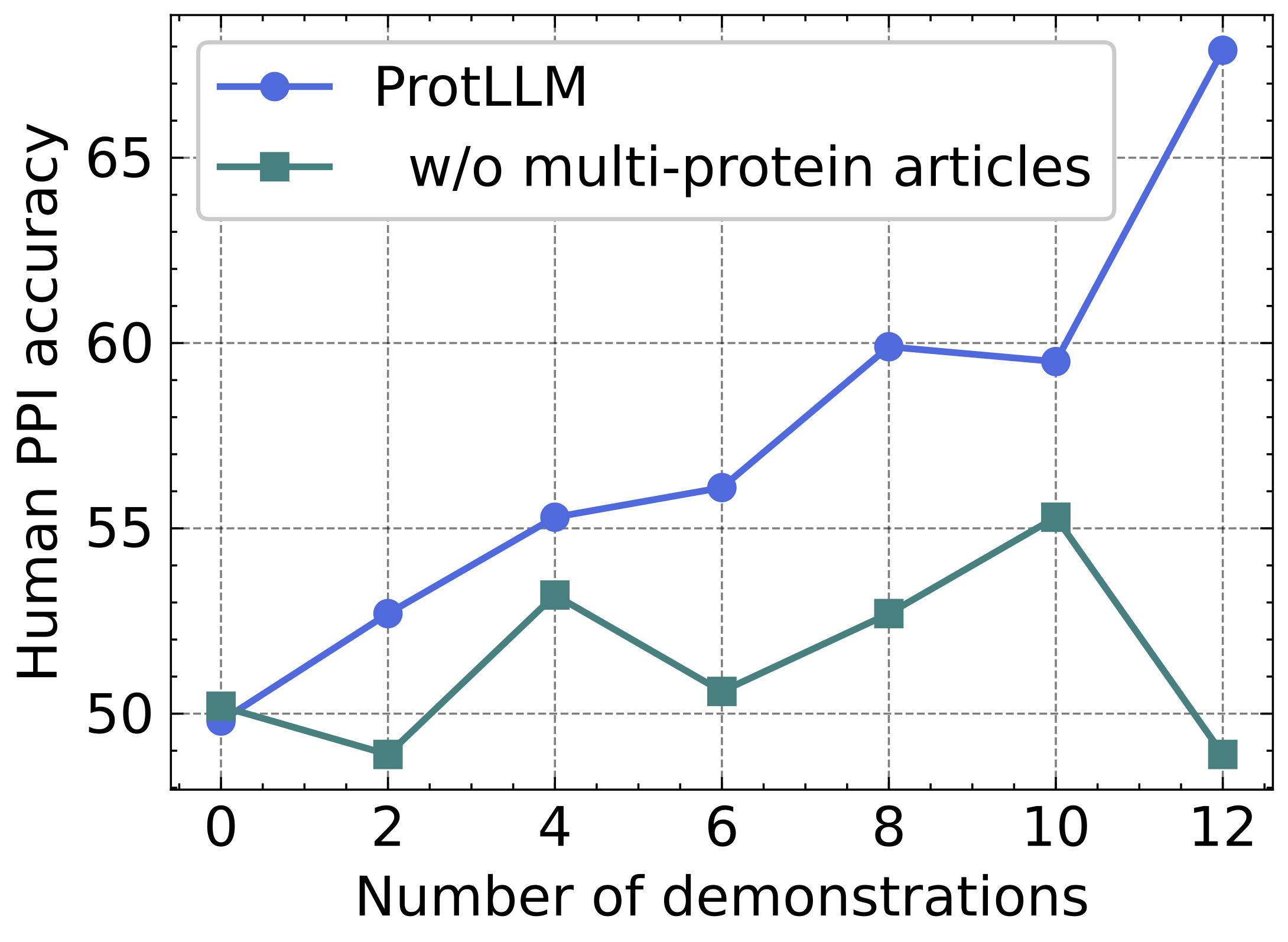
In-context learning results on human PPI.
This experiment aims to study the capability of ProtLLM to retrieve functional proteins based on text prompts and demonstrations. For this purpose, we apply ProtLLM to enzyme mining, which is a critical stage in enzyme and metabolic engineering pipelines. In this experiment, we evaluate our model on mining carboxylate reductases that transform various ketoacids into their corresponding aldehydes. Four ketoacid reactants, i.e., 2-ketoisovaleric acid (IsoC5), pyruvic acid (C3), 2-ketovaleric acid (C5), and 2-ketooctanoic acid (C8) are employed for evaluation. Using a reported enzyme for IsoC5, ketoisovalerate decarboxylase (KIVD), as the query, we first search for a pool of enzyme candidates by BLASTp, where the pools with the size of 500 and 1000 are respectively tested. We then leverage ProtLLM to retrieve active enzymes from the pool for each reactant in two modes.
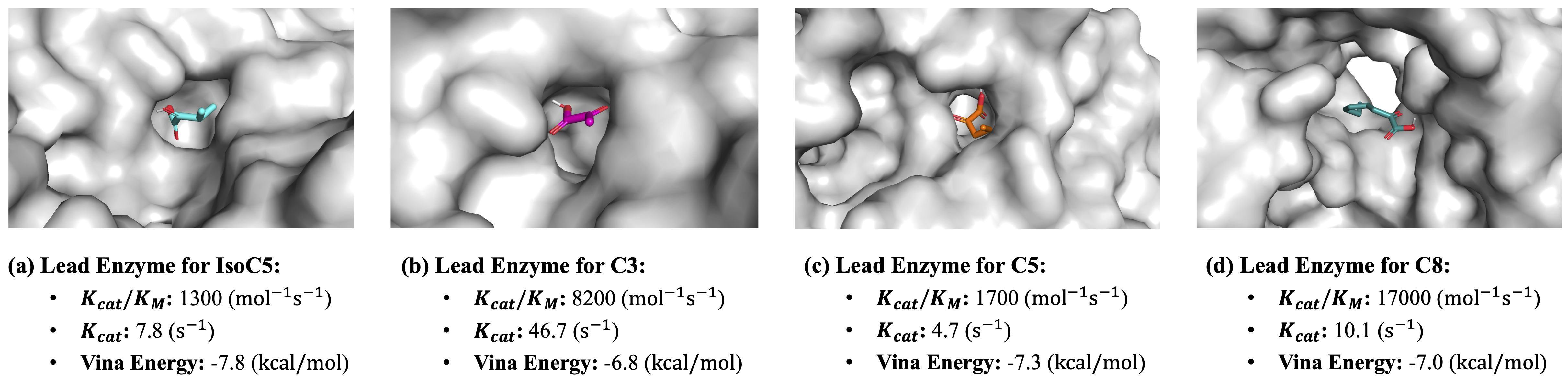
Top-1 enzyme mining results based on ProtLLM retrieval and AutoDock Vina post-screening. \(K_{cat}/K_M\) and \(K_{cat}\) measure enzyme activity (higher the better). Vina energy measures binding affinity (lower the better).
In the following table, we report the recall of active enzymes at top 10, 20, and 50 ranked candidates. It is observed that in-context learning outperforms zero-shot retrieval on 18 out of 24 metrics, which verifies that ProtLLM can learn from a few demonstrations and improve its enzyme mining performance based on such knowledge. To study the top-ranked enzymes by ProtLLM more in depth, we employ AutoDock Vina to further screen the top-20 enzymes found by in-context learning and pick the one with the lowest Vina energy for visualization. As shown in the figure above, the lead enzymes selected in this way are all with good properties, possessing high enzyme activity (i.e., high \(K_{cat} / K_M\) and \(K_{cat}\) values) and low binding energy measured by AutoDock Vina. These results altogether prove the effectiveness of ProtLLM on enzyme mining.
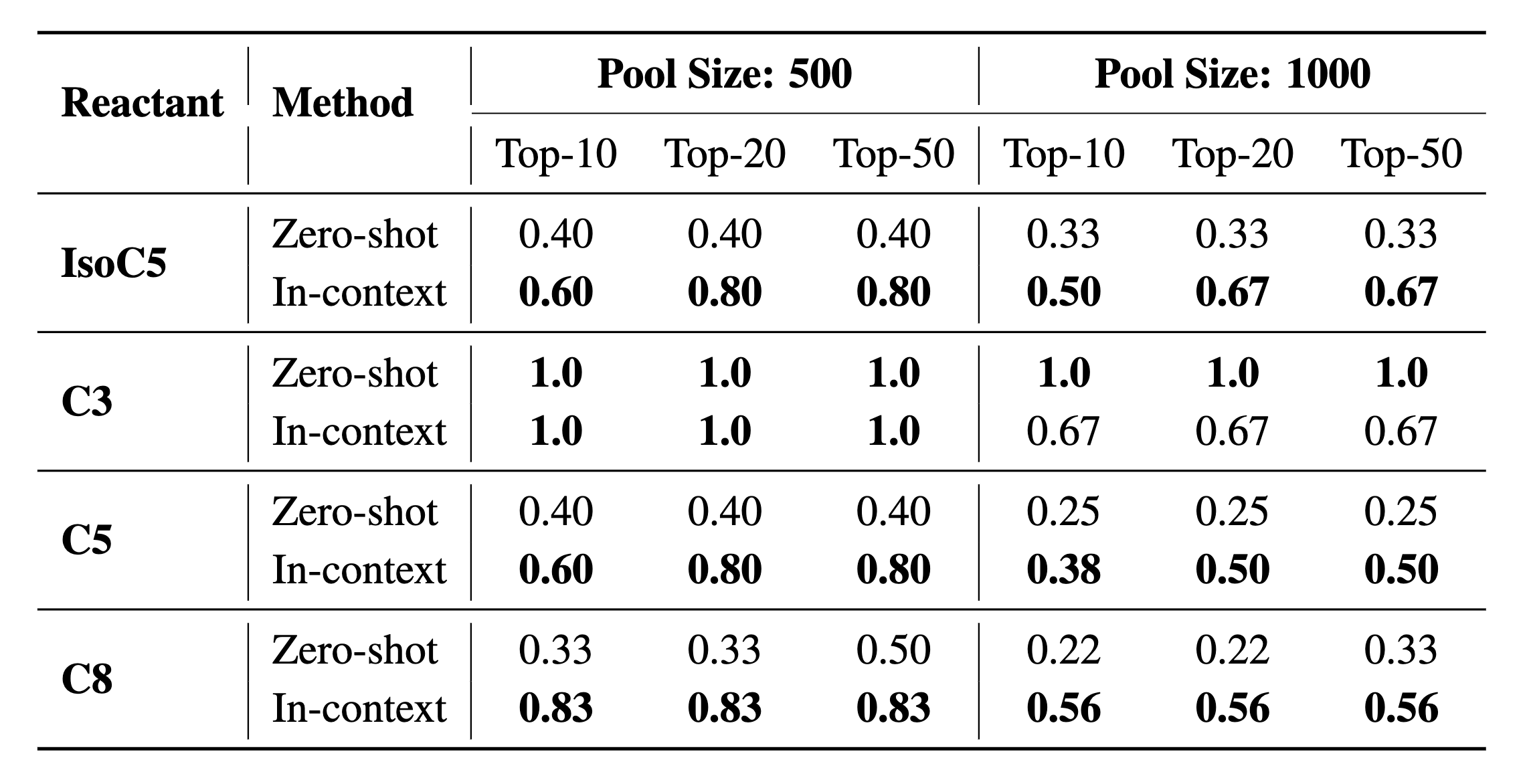
Performance comparisons between zero-shot retrieval and in-context learning on enzyme mining. Top-10, 20 and 50 Recall are reported.
We give special thanks to Jianan Zhao, Zuobai Zhang, Meng Qu, and Kunlun Zhu for their helpful discussions and comments.
@article{zhuo2024protllm,
title={ProtLLM: An Interleaved Protein-Language LLM with Protein-as-Word Pre-Training},
author={Le Zhuo and Zewen Chi and Minghao Xu and Heyan Huang and Heqi Zheng and Conghui He and Xian-Ling Mao and Wentao Zhang},
journal={arXiv preprint arXiv:2403.079205},
year={2024}
}